
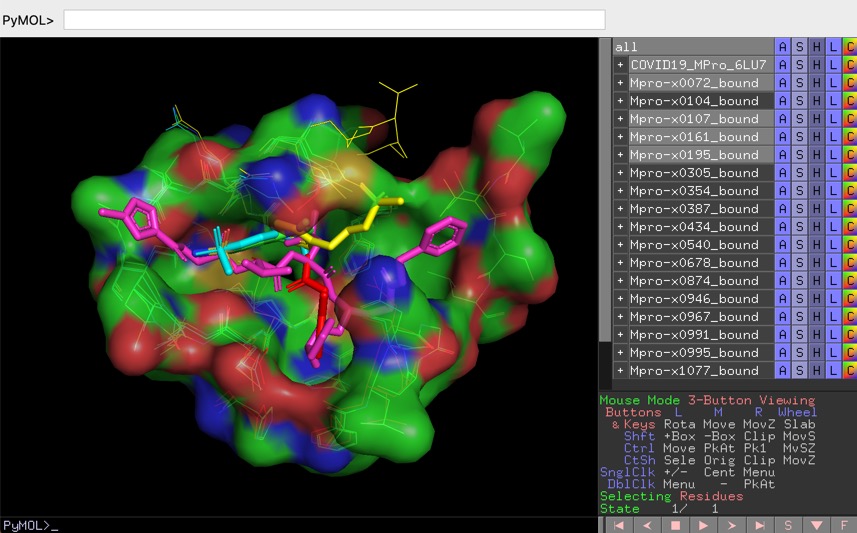
You should separate the ligand and the protein file of this pdb file. We will continue working on the 2amb structure we used at the end of the pymol tutorial. Then click on the AutoDockTools 1.5.0 icon to start the interface. Go to the Kuabuntu menu and search for the MGLtools. Probably less visually attractive than pymol, the autodock interface ( called ADT) offers pretty much the same standard functionalities with the advantage of an optimized environment to preparing autodock inputs. To prepare our files, we will use the visual interface available for Autodock that allow to prepare, study and analysis dockings. The different steps for the initial study as well as the possible extension of the work are summarize below.Īutodock scoring function is applied using an adapted AMBER force field therefore the atoms of the protein and the ligand have to be set up in accordance with this ff. In this tutorial, you will be introduced the docking approaches by undertaking undertaking on your own some docking experiments on the androgen binding receptor seen in the tutorial I. The actual version of the program (the version we will use here) is the version 4.0 which provides with important new features for docking prospect like protein residue flexibility and high quality scoring functions. Some of the algorithms detailed during the theoretical courses are available in Autodock.
Pymol tutorial how to show ligand interactions download#
You can download all the files of this tutorial hereĪutodock is a flexible ligand-protein docking program which basically runs as a two steps procedure: the calculation of the map of interactions of the binding site with some general atom types (performed with autogrid) and the posing of the lligand respecting this map of interaction (performed with autodock).


 0 kommentar(er)
0 kommentar(er)
